
Contributions
Abstract: PB2308
Type: Publication Only
Background
Bone marrow (BM) aspiration is a standard diagnostic and evaluation tool utilized by physicians together with other exams to make clinical decisions for all major hematologic diseases. The morphology report includes many important morphological features of the specimens. However, these findings are often documented in an unstructured free-text format, making it extremely difficult to extract and compare information from different reports. Recently, successful applications of natural language processing (NLP) techniques for automatic categorizations of findings on pathology or electronic medical reports have been shown for diseases such as breast cancer diagnoses.
Aims
We aim to develop NLP algorithms to automatically extract key clinical information to support physicians in the clinical decision and further research for hematologic malignancies.
Methods
A total of 12549 free-text BM morphology reports dated from 2007 to 2016 were collected from the National Taiwan University Hospital. Each report consists of information on the following key clinical classifications in a free-text format: specimen quality, disease types and disease status. We first achieved word normalization of these reports through a series lexical pre-processing using Wordnet lexicon corpus. We then derived a document vector as a summary representation of each report. This vector includes a fixed dimension equals to the sum total of unique uni- bi- tri-gram word count in our BM dataset, and each ith element reflects the importance weighting for each word in the given report. The importance weighting is computed based on term-frequency inverse-document frequency (tf(t,d)*idf(d,t)). Term frequency, tf(t,d), is the number of occurrences of ith word in the dth report. And, idf(d,t)=log(n/(df(d,t)+1) where df(d,t) indicates the total number of report containing ith word and n is the total number of reports. We finally trained machine learning algorithms to automatically learn to extract key clinical information using class-balanced support vector machine (SVM) with this tf-idf document vectors.
We evaluated our approach using a three-fold cross-validation. That is, for each fold, 9790 reports were used as training set and the remaining 2759 reports were used as the blind testing set and we iterated this for three times. The accuracy was measured by comparing the concordance rate between the algorithm categorization and the expert categorization on the testing set.
Results
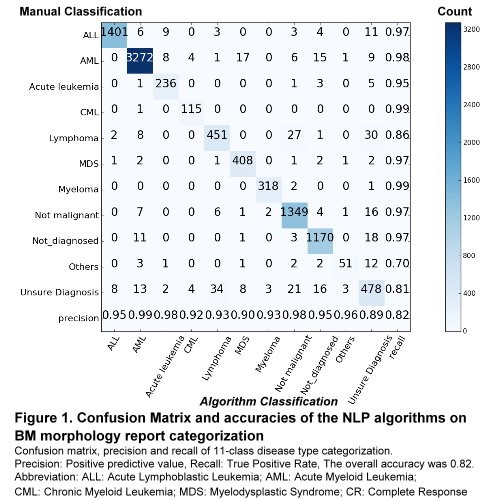
All three NLP algorithms achieved high categorization performance. The disease type classification achieved 0.95 accuracies in 9 out of 11 types of categories (Figure 1). In addition, the other two algorithms achieved overall 0.98 accuracies in specimen quality classification and 0.95 in disease status classification (data not shown). Among the 11 disease type classification tasks, the algorithm achieved accuracy lower than 0.95 in 3 categories: “Lymphoma”, “Other” and “Unsure Diagnosis”. The relatively lower accuracy of the ”Lymphoma” class may be attributed to the fact that it was used to include all subtypes of lymphoma. Similarly, the “Others” class was used as the “garbage” class where multiple diagnostics decision other than malignancies could be made, and the “Unsure Diagnosis” reflect uncertain clinical diagnostic decisions.
Conclusion
Our NLP algorithms could efficiently and accurately extract important information with high accuracy from a large scale free-text bone marrow morphology reports. Further research is needed to evaluate the algorithm performance on reports from other centers.
Session topic: 16. Myeloproliferative neoplasms - Clinical
Keyword(s): Bone Marrow, Hematological malignancy, Pathology
Abstract: PB2308
Type: Publication Only
Background
Bone marrow (BM) aspiration is a standard diagnostic and evaluation tool utilized by physicians together with other exams to make clinical decisions for all major hematologic diseases. The morphology report includes many important morphological features of the specimens. However, these findings are often documented in an unstructured free-text format, making it extremely difficult to extract and compare information from different reports. Recently, successful applications of natural language processing (NLP) techniques for automatic categorizations of findings on pathology or electronic medical reports have been shown for diseases such as breast cancer diagnoses.
Aims
We aim to develop NLP algorithms to automatically extract key clinical information to support physicians in the clinical decision and further research for hematologic malignancies.
Methods
A total of 12549 free-text BM morphology reports dated from 2007 to 2016 were collected from the National Taiwan University Hospital. Each report consists of information on the following key clinical classifications in a free-text format: specimen quality, disease types and disease status. We first achieved word normalization of these reports through a series lexical pre-processing using Wordnet lexicon corpus. We then derived a document vector as a summary representation of each report. This vector includes a fixed dimension equals to the sum total of unique uni- bi- tri-gram word count in our BM dataset, and each ith element reflects the importance weighting for each word in the given report. The importance weighting is computed based on term-frequency inverse-document frequency (tf(t,d)*idf(d,t)). Term frequency, tf(t,d), is the number of occurrences of ith word in the dth report. And, idf(d,t)=log(n/(df(d,t)+1) where df(d,t) indicates the total number of report containing ith word and n is the total number of reports. We finally trained machine learning algorithms to automatically learn to extract key clinical information using class-balanced support vector machine (SVM) with this tf-idf document vectors.
We evaluated our approach using a three-fold cross-validation. That is, for each fold, 9790 reports were used as training set and the remaining 2759 reports were used as the blind testing set and we iterated this for three times. The accuracy was measured by comparing the concordance rate between the algorithm categorization and the expert categorization on the testing set.
Results
All three NLP algorithms achieved high categorization performance. The disease type classification achieved 0.95 accuracies in 9 out of 11 types of categories (Figure 1). In addition, the other two algorithms achieved overall 0.98 accuracies in specimen quality classification and 0.95 in disease status classification (data not shown). Among the 11 disease type classification tasks, the algorithm achieved accuracy lower than 0.95 in 3 categories: “Lymphoma”, “Other” and “Unsure Diagnosis”. The relatively lower accuracy of the ”Lymphoma” class may be attributed to the fact that it was used to include all subtypes of lymphoma. Similarly, the “Others” class was used as the “garbage” class where multiple diagnostics decision other than malignancies could be made, and the “Unsure Diagnosis” reflect uncertain clinical diagnostic decisions.

Conclusion
Our NLP algorithms could efficiently and accurately extract important information with high accuracy from a large scale free-text bone marrow morphology reports. Further research is needed to evaluate the algorithm performance on reports from other centers.
Session topic: 16. Myeloproliferative neoplasms - Clinical
Keyword(s): Bone Marrow, Hematological malignancy, Pathology


