
Contributions
Abstract: S867
Type: Oral Presentation
Presentation during EHA23: On Saturday, June 16, 2018 from 16:00 - 16:15
Location: Room K1
Background
Trisomy 12 (Tri12) chronic lymphocytic leukemia (CLL) identifies a cytogenetic subset with a peculiar clinical behavior (Bulian et al, Haematologica, 2017) and specific biological features, including high frequency of stabilizing NOTCH1 mutations (NOTCH1-mut) (Rossi et al, Blood, 2013). Moreover, KRAS and its transcriptional regulator HNRNPA1, key components of Ras-Raf-MEK-ERK pathway are both hosted in the trisomic chromosome 12.
Aims
To investigate the role of KRAS expression and/or activation in Tri12 and NOTCH1-mut CLL.
Methods
Tri12 was assessed by FISH. NOTCH1 and KRAS mutations were assessed by either Sanger or NGS. QRT-PCR and western blot were employed to evaluate HNRNPA1 and KRAS expression and down-stream signaling in primary CLL or CLL cell line models. Active GTP-bound RAS pull-down was performed by Raf1 RBD agarose beads assay. CI and MEC-1 cell lines were used as human in vitro model of Tri12 NOTCH1-mut CLL and non-Tri12 NOTCH1-wild type (wt) CLL, respectively. Mann-Whitney test, unpaired t-test or Chi-Square test were used to compare differences between groups.
Results
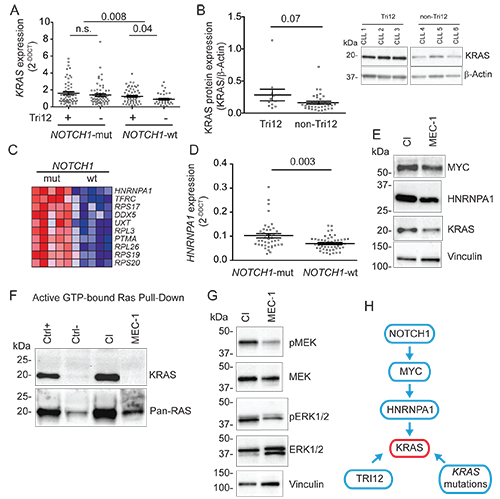
KRAS transcript was analyzed in 215 cases purposely enriched in Tri12 (118) and NOTCH1-mut (121). As shown in Fig. A, Tri12 were characterized by higher KRAS level in the context of NOTCH1-wt CLL (p=0.04); conversely, no differences in KRAS expression were found between Tri12 and non-Tri12 in the context of NOTCH1-mut CLL cases, which however expressed higher KRAS transcript compared to NOTCH1-wt CLL (p=0.008). Consistently, KRAS protein level was higher in Tri12 vs. non-Tri12 in the context of NOTCH1-wt CLL (Fig. B). A gene expression profiling identified HNRNPA1 as the top ranked gene among MYC target genes upregulated in NOTCH1-mut vs. NOTCH1-wt CLL. Accordingly, higher HNRNPA1 levels were found by QRT-PCR in additional 41 NOTCH1-mut vs. 47 NOTCH1-wt CLL samples (p=0.003, Fig. C,D) without differences between Tri12 (42) and non-Tri12 (46) CLL. In line with a KRAS transcriptional activation mediated by NOTCH1-MYC-HNRNPA1 axis, higher MYC, HNRNPA1 and KRAS protein levels were found in the Tri12 NOTCH1-mut CI cell line when compared to the non-Tri12 NOTCH1-wt MEC-1 cells. Furthermore, higher levels of active GTP-bound KRAS and higher phosphorylation levels of MEK and ERK were found in CI cells (Fig. E, F, G), supporting the hypothesis of sustained Ras-Raf-MEK-ERK signaling in Tri12 and NOTCH1-mut CLL. Finally, analysis of KRAS genomic aberrations revealed higher mutations incidence in Tri12 CLL (10/77, 13%) when compared to non-Tri12 CLL (2/68, 2.9%, p=0.03). Altogether, these data foster the hypothesis of multiple mechanisms of KRAS overexpression/activation occurring in Tri12 CLL via the NOTCH1-MYC-HNRNPA1 axis in NOTCH1-mut cases, or due to a higher incidence of KRAS mutations or an overexpression of KRAS by the supernumerary chromosome 12(Fig. H).
Conclusion
Our data, by describing a synergism between Tri12, NOTCH1-mut and KRAS-mut in boosting KRAS expression and activity in CLL, indicate the Tri12 subset as particularly addicted to the Ras-Raf-MEK-ERK signaling pathway and likely to benefit of ERK/MEK inhibitors, as recently emphasized (Dietrich et al, J Clin Invest, 2018).
Session topic: 5. Chronic lymphocytic leukemia and related disorders – Biology & Translational Research
Keyword(s): Chronic Lymphocytic Leukemia, Notch signaling, Ras
Abstract: S867
Type: Oral Presentation
Presentation during EHA23: On Saturday, June 16, 2018 from 16:00 - 16:15
Location: Room K1
Background
Trisomy 12 (Tri12) chronic lymphocytic leukemia (CLL) identifies a cytogenetic subset with a peculiar clinical behavior (Bulian et al, Haematologica, 2017) and specific biological features, including high frequency of stabilizing NOTCH1 mutations (NOTCH1-mut) (Rossi et al, Blood, 2013). Moreover, KRAS and its transcriptional regulator HNRNPA1, key components of Ras-Raf-MEK-ERK pathway are both hosted in the trisomic chromosome 12.
Aims
To investigate the role of KRAS expression and/or activation in Tri12 and NOTCH1-mut CLL.
Methods
Tri12 was assessed by FISH. NOTCH1 and KRAS mutations were assessed by either Sanger or NGS. QRT-PCR and western blot were employed to evaluate HNRNPA1 and KRAS expression and down-stream signaling in primary CLL or CLL cell line models. Active GTP-bound RAS pull-down was performed by Raf1 RBD agarose beads assay. CI and MEC-1 cell lines were used as human in vitro model of Tri12 NOTCH1-mut CLL and non-Tri12 NOTCH1-wild type (wt) CLL, respectively. Mann-Whitney test, unpaired t-test or Chi-Square test were used to compare differences between groups.
Results
KRAS transcript was analyzed in 215 cases purposely enriched in Tri12 (118) and NOTCH1-mut (121). As shown in Fig. A, Tri12 were characterized by higher KRAS level in the context of NOTCH1-wt CLL (p=0.04); conversely, no differences in KRAS expression were found between Tri12 and non-Tri12 in the context of NOTCH1-mut CLL cases, which however expressed higher KRAS transcript compared to NOTCH1-wt CLL (p=0.008). Consistently, KRAS protein level was higher in Tri12 vs. non-Tri12 in the context of NOTCH1-wt CLL (Fig. B). A gene expression profiling identified HNRNPA1 as the top ranked gene among MYC target genes upregulated in NOTCH1-mut vs. NOTCH1-wt CLL. Accordingly, higher HNRNPA1 levels were found by QRT-PCR in additional 41 NOTCH1-mut vs. 47 NOTCH1-wt CLL samples (p=0.003, Fig. C,D) without differences between Tri12 (42) and non-Tri12 (46) CLL. In line with a KRAS transcriptional activation mediated by NOTCH1-MYC-HNRNPA1 axis, higher MYC, HNRNPA1 and KRAS protein levels were found in the Tri12 NOTCH1-mut CI cell line when compared to the non-Tri12 NOTCH1-wt MEC-1 cells. Furthermore, higher levels of active GTP-bound KRAS and higher phosphorylation levels of MEK and ERK were found in CI cells (Fig. E, F, G), supporting the hypothesis of sustained Ras-Raf-MEK-ERK signaling in Tri12 and NOTCH1-mut CLL. Finally, analysis of KRAS genomic aberrations revealed higher mutations incidence in Tri12 CLL (10/77, 13%) when compared to non-Tri12 CLL (2/68, 2.9%, p=0.03). Altogether, these data foster the hypothesis of multiple mechanisms of KRAS overexpression/activation occurring in Tri12 CLL via the NOTCH1-MYC-HNRNPA1 axis in NOTCH1-mut cases, or due to a higher incidence of KRAS mutations or an overexpression of KRAS by the supernumerary chromosome 12(Fig. H).

Conclusion
Our data, by describing a synergism between Tri12, NOTCH1-mut and KRAS-mut in boosting KRAS expression and activity in CLL, indicate the Tri12 subset as particularly addicted to the Ras-Raf-MEK-ERK signaling pathway and likely to benefit of ERK/MEK inhibitors, as recently emphasized (Dietrich et al, J Clin Invest, 2018).
Session topic: 5. Chronic lymphocytic leukemia and related disorders – Biology & Translational Research
Keyword(s): Chronic Lymphocytic Leukemia, Notch signaling, Ras


