
Contributions
Abstract: S830
Type: Oral Presentation
Presentation during EHA23: On Saturday, June 16, 2018 from 12:15 - 12:30
Location: Room A7
Background
Gene expression profiling has enabled the identification of three distinct DLBCL subtypes: ABC, GCB and PMBL. More recently, techniques such as Next Generation Sequencing (NGS) and Copy Number Variation (CNV) analysis have led to an increasingly detailed characterization of the genomic profiles of DLBCL.
Aims
The aim of this study was to perform a fully integrated analysis of mutational, genomic, and expression profiles of DLBCL to refine disease subtypes using molecular tools.
Methods
215 patients with de novo DLBCL from the prospective, multicenter and randomized LNH-03B LYSA clinical trials were included. Gene expression profiling (GEP) was obtained with HGU133+2.0 Affymetrix GeneChip arrays. Mutational profiles were established by Lymphopanel NGS, targeting 34 key genes (Dubois et al, CCR 2016). CNV were analyzed by array Comparative Genomic Hybridization. FISH was performed to identify BCL2, BCL6 and MYC rearrangements. IHC was performed on CD10, BCL6, MUM1, BCL2, FOXP1, IgM and MYC. We applied unsupervised independent component analysis (ICA) to GEP data and identified 38 gene signatures (components) reflecting transcriptomic variability across our DLBCL cohort. SignatureDB and MSigDB databases were used to interpret the biological function of each component.
Results
Many of the components were closely related to well-known DLBCL features such as cell of origin, stromal and MYC signatures, as evidenced by gene set overrepresentation analyses. Correlation of the components’ expression with gene mutational status, CNV data, IHC and FISH identified interesting associations.
Stromal components correlated positively with B2M and TNFAIP3 mutations (FDR≈0.02) and negatively with CD79B mutations (FDR≈0.008). The GCB/ABC signature correlated positively with EZH2, BCL2, STAT6 and GNA13 mutations (FDR <10-3 -0.06) and negatively with CD79B and PIM1 mutations (FDR 0.005 - 0.06).
PMBL and interferon components shared numerous correlations with mutations including SOCS1, B2M, ITPKB, STAT6, CD58, XPO1 and TNFAIP3 (FDR <10-3 - 0.04). Both components were also linked to gain/amplification of the 9p24 locus containing PDL1/PDL2. Interestingly, PMBL with high interferon component expression also presented low T cell component expression, in keeping with the negative regulation of T lymphocytes by PDL1/2 during the immune response. ICA also identified variability among the GCB subgroup, delineating proliferation-high / T cell-low GCB patients versus GCB patients with the opposite phenotype.
The Myc component was strongly linked to FOXP1, MUM1, IgM and MYC IHC expression, as well as to MYC rearrangement, showcasing a poor prognosis non-GCB population.
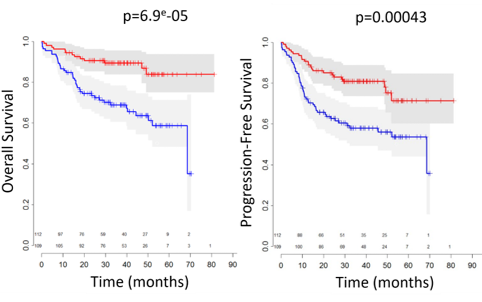
Stromal components were strongly correlated with better OS (p<10-4 - p<0.05) and PFS (p<10-3) (Figure). Another component correlated with MYD88 and CD79B mutations as well as gain of 19q13 locus was found to be correlated with poor OS and PFS (p=0.01): gain of 19q13 locus has previously been shown to be linked to reduced event-free survival.
Conclusion
ICA is an unsupervised approach, which is sensitive and efficient for the analysis of DLBCL expression profiles. We characterized components linked to gene mutational status, to the presence of CNV, to IHC and FISH results and, notably, to patient outcome. The results of this type of integrated analysis should enable a global and multi-level view of DLBCL, as well as improve our understanding of DLBCL subgroups.
Session topic: 19. Non-Hodgkin lymphoma Biology & Translational Research
Keyword(s): DLBCL, Expression profiling, Genomics, mutation analysis
Abstract: S830
Type: Oral Presentation
Presentation during EHA23: On Saturday, June 16, 2018 from 12:15 - 12:30
Location: Room A7
Background
Gene expression profiling has enabled the identification of three distinct DLBCL subtypes: ABC, GCB and PMBL. More recently, techniques such as Next Generation Sequencing (NGS) and Copy Number Variation (CNV) analysis have led to an increasingly detailed characterization of the genomic profiles of DLBCL.
Aims
The aim of this study was to perform a fully integrated analysis of mutational, genomic, and expression profiles of DLBCL to refine disease subtypes using molecular tools.
Methods
215 patients with de novo DLBCL from the prospective, multicenter and randomized LNH-03B LYSA clinical trials were included. Gene expression profiling (GEP) was obtained with HGU133+2.0 Affymetrix GeneChip arrays. Mutational profiles were established by Lymphopanel NGS, targeting 34 key genes (Dubois et al, CCR 2016). CNV were analyzed by array Comparative Genomic Hybridization. FISH was performed to identify BCL2, BCL6 and MYC rearrangements. IHC was performed on CD10, BCL6, MUM1, BCL2, FOXP1, IgM and MYC. We applied unsupervised independent component analysis (ICA) to GEP data and identified 38 gene signatures (components) reflecting transcriptomic variability across our DLBCL cohort. SignatureDB and MSigDB databases were used to interpret the biological function of each component.
Results
Many of the components were closely related to well-known DLBCL features such as cell of origin, stromal and MYC signatures, as evidenced by gene set overrepresentation analyses. Correlation of the components’ expression with gene mutational status, CNV data, IHC and FISH identified interesting associations.
Stromal components correlated positively with B2M and TNFAIP3 mutations (FDR≈0.02) and negatively with CD79B mutations (FDR≈0.008). The GCB/ABC signature correlated positively with EZH2, BCL2, STAT6 and GNA13 mutations (FDR <10-3 -0.06) and negatively with CD79B and PIM1 mutations (FDR 0.005 - 0.06).
PMBL and interferon components shared numerous correlations with mutations including SOCS1, B2M, ITPKB, STAT6, CD58, XPO1 and TNFAIP3 (FDR <10-3 - 0.04). Both components were also linked to gain/amplification of the 9p24 locus containing PDL1/PDL2. Interestingly, PMBL with high interferon component expression also presented low T cell component expression, in keeping with the negative regulation of T lymphocytes by PDL1/2 during the immune response. ICA also identified variability among the GCB subgroup, delineating proliferation-high / T cell-low GCB patients versus GCB patients with the opposite phenotype.
The Myc component was strongly linked to FOXP1, MUM1, IgM and MYC IHC expression, as well as to MYC rearrangement, showcasing a poor prognosis non-GCB population.
Stromal components were strongly correlated with better OS (p<10-4 - p<0.05) and PFS (p<10-3) (Figure). Another component correlated with MYD88 and CD79B mutations as well as gain of 19q13 locus was found to be correlated with poor OS and PFS (p=0.01): gain of 19q13 locus has previously been shown to be linked to reduced event-free survival.

Conclusion
ICA is an unsupervised approach, which is sensitive and efficient for the analysis of DLBCL expression profiles. We characterized components linked to gene mutational status, to the presence of CNV, to IHC and FISH results and, notably, to patient outcome. The results of this type of integrated analysis should enable a global and multi-level view of DLBCL, as well as improve our understanding of DLBCL subgroups.
Session topic: 19. Non-Hodgkin lymphoma Biology & Translational Research
Keyword(s): DLBCL, Expression profiling, Genomics, mutation analysis


