
Contributions
Abstract: S494
Type: Oral Presentation
Presentation during EHA22: On Saturday, June 24, 2017 from 16:45 - 17:00
Location: Room N103
Background
Aims
Methods
We integrated molecular data with available prognostic factors in patients undergoing alloHCT for MDS and sAML to evaluate their impact on prognosis. 304 patients with MDS or sAML who underwent alloHCT were sequenced for mutations in 54 genes. We used a Cox multivariate model and competing risk analysis with internal and cross validation to identify factors prognostic of overall survival (OS), cumulative incidence of relapse (CIR) and non-relapse mortality (NRM).
Results
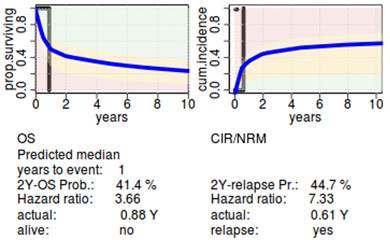
Conclusion
Session topic: 22. Stem cell transplantation - Clinical
Keyword(s): AML, Transplant, prognosis, MDS
Abstract: S494
Type: Oral Presentation
Presentation during EHA22: On Saturday, June 24, 2017 from 16:45 - 17:00
Location: Room N103
Background
Aims
Methods
We integrated molecular data with available prognostic factors in patients undergoing alloHCT for MDS and sAML to evaluate their impact on prognosis. 304 patients with MDS or sAML who underwent alloHCT were sequenced for mutations in 54 genes. We used a Cox multivariate model and competing risk analysis with internal and cross validation to identify factors prognostic of overall survival (OS), cumulative incidence of relapse (CIR) and non-relapse mortality (NRM).
Results

Conclusion
Session topic: 22. Stem cell transplantation - Clinical
Keyword(s): AML, Transplant, prognosis, MDS


