COMPARISON OF GENOMIC DNA AND REVERSE TRANSCRIPTASE Q-PCR FOR THE MONITORING OF FIRST-LINE IMATINIB TREATMENT: AN ALLG CML9 SUB-STUDY
(Abstract release date: 05/18/17)
EHA Library. S. Pagani I. 06/24/17; 181771; S484

Dr. Ilaria S. Pagani
Contributions
Contributions
Abstract
Abstract: S484
Type: Oral Presentation
Presentation during EHA22: On Saturday, June 24, 2017 from 16:45 - 17:00
Location: Room N101
Background
Real-time reverse transcriptase quantitative PCR (RQ-PCR) for BCR-ABL1 mRNA is widely used for the monitoring of chronic myeloid leukaemia (CML). Pre-analytical factors, such as the rate of degradation of the target mRNA, and methodological factors, such as the choice of control gene, may influence the final result. In contrast the genomic DNA is stable, and the number of copies of BCR-ABL1 DNA is directly proportional to the number of CML cells. Measuring both DNA and RNA may enable us to understand the contribution of expression and cell number to the RQ-PCR response.
Aims
To compare BCR-ABL1 DNA Q-PCR and routine RQ-PCR monitoring of CML.
Methods
Fifty-nine newly diagnosed chronic phase CML patients from the ALLG CML9 (TIDEL II) trial were included in this sub-study. Samples were tested prior to commencing TKI treatment (baseline), at 1, 2, and 3 months, and every 3 months to 24 months (total 568 samples). Since we wanted to compare the sensitivity of the Q-PCR methods we selected 19 patients who had achieved undetectable minimal residual disease (UMRD) by RQ-PCR within 24 months, and an additional 40 patients unselected for response. RQ-PCR results were expressed on the International Scale (IS), whereas DNA results were expressed relative to the individual patient’s baseline. Quantification of BCR-ABL1 DNA was performed using GUSB as the control gene by real-time Q-PCR (n=40) or by digital PCR (dPCR, n=19) using the Fluidigm BioMark HD System. The mean detection limit of RQ-PCR was 4.5-log, and 5.4-log for DNA methods.
Results
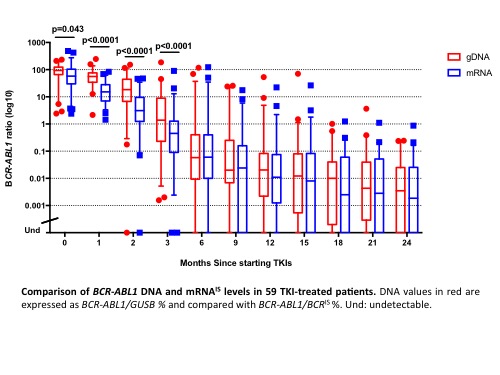
We first demonstrated that DNA dPCR and real-time Q-PCR gave comparable results: 45 samples from 6 patients were quantified by both methods and logarithmically transformed. The mean bias was -0.15 (1.4-fold) with 95% limits of agreement ranging from -1.19 to 0.88. Subsequently, DNA and mRNA values were compared in paired samples. The median BCR-ABL1IS at baseline was 58% (range, 2.4% - 487%) versus 93% by DNA methods (range, 2.4% - 235%). Interestingly, BCR-ABL1 DNA was significantly higher than mRNA at 1, 2, and 3 months (Figure). There was good agreement between positive results from 6 months of TKI therapy onwards (mean bias -0.02; 95% limits of agreement from -1.15 to 1.11). Comparing the limit of detection, BCR-ABL1 DNA was detectable in 60/148 (41%) samples with undetectable mRNA (median 0.002%, range 0.0003-0.07%). Finally, 88% (15/17) of patients with e14a2 transcripts achieved MMR by 12 months, in comparison with 63% (20/32) of patients with e13a2 transcripts. BCR-ABL1 mRNA expression levels were significantly lower in e13a2 patients than in e14a2 patients in all follow-up samples analysed (ratio of BCR-ABL1 mRNA:DNA for e13a2 0.44 vs e14a2 0.57; p=0.016).
Conclusion
In the first 1-3 months BCR-ABL1 mRNA fell more rapidly than DNA, likely reflecting the time taken for normal haematopoietic cells to recover. At later time-points there was good agreement between methods, indicating that later reduction in BCR-ABL1IS is closely related to depletion of leukaemic cells. Normalised to BCR-ABL1 DNA the expression of e13a2 BCR-ABL1 mRNA was lower than that of e14a2, an observation that requires confirmation. DNA methods were more sensitive: following the achievement of UMRD by RQ-PCR patients could, on average, be monitored by DNA Q-PCR for an additional 5 months.
Session topic: 8. Chronic myeloid leukemia - Clinical
Keyword(s): BCR-ABL, Molecular response, Minimal residual disease (MRD), Chronic myeloid leukemia
Abstract: S484
Type: Oral Presentation
Presentation during EHA22: On Saturday, June 24, 2017 from 16:45 - 17:00
Location: Room N101
Background
Real-time reverse transcriptase quantitative PCR (RQ-PCR) for BCR-ABL1 mRNA is widely used for the monitoring of chronic myeloid leukaemia (CML). Pre-analytical factors, such as the rate of degradation of the target mRNA, and methodological factors, such as the choice of control gene, may influence the final result. In contrast the genomic DNA is stable, and the number of copies of BCR-ABL1 DNA is directly proportional to the number of CML cells. Measuring both DNA and RNA may enable us to understand the contribution of expression and cell number to the RQ-PCR response.
Aims
To compare BCR-ABL1 DNA Q-PCR and routine RQ-PCR monitoring of CML.
Methods
Fifty-nine newly diagnosed chronic phase CML patients from the ALLG CML9 (TIDEL II) trial were included in this sub-study. Samples were tested prior to commencing TKI treatment (baseline), at 1, 2, and 3 months, and every 3 months to 24 months (total 568 samples). Since we wanted to compare the sensitivity of the Q-PCR methods we selected 19 patients who had achieved undetectable minimal residual disease (UMRD) by RQ-PCR within 24 months, and an additional 40 patients unselected for response. RQ-PCR results were expressed on the International Scale (IS), whereas DNA results were expressed relative to the individual patient’s baseline. Quantification of BCR-ABL1 DNA was performed using GUSB as the control gene by real-time Q-PCR (n=40) or by digital PCR (dPCR, n=19) using the Fluidigm BioMark HD System. The mean detection limit of RQ-PCR was 4.5-log, and 5.4-log for DNA methods.
Results
We first demonstrated that DNA dPCR and real-time Q-PCR gave comparable results: 45 samples from 6 patients were quantified by both methods and logarithmically transformed. The mean bias was -0.15 (1.4-fold) with 95% limits of agreement ranging from -1.19 to 0.88. Subsequently, DNA and mRNA values were compared in paired samples. The median BCR-ABL1IS at baseline was 58% (range, 2.4% - 487%) versus 93% by DNA methods (range, 2.4% - 235%). Interestingly, BCR-ABL1 DNA was significantly higher than mRNA at 1, 2, and 3 months (Figure). There was good agreement between positive results from 6 months of TKI therapy onwards (mean bias -0.02; 95% limits of agreement from -1.15 to 1.11). Comparing the limit of detection, BCR-ABL1 DNA was detectable in 60/148 (41%) samples with undetectable mRNA (median 0.002%, range 0.0003-0.07%). Finally, 88% (15/17) of patients with e14a2 transcripts achieved MMR by 12 months, in comparison with 63% (20/32) of patients with e13a2 transcripts. BCR-ABL1 mRNA expression levels were significantly lower in e13a2 patients than in e14a2 patients in all follow-up samples analysed (ratio of BCR-ABL1 mRNA:DNA for e13a2 0.44 vs e14a2 0.57; p=0.016).

Conclusion
In the first 1-3 months BCR-ABL1 mRNA fell more rapidly than DNA, likely reflecting the time taken for normal haematopoietic cells to recover. At later time-points there was good agreement between methods, indicating that later reduction in BCR-ABL1IS is closely related to depletion of leukaemic cells. Normalised to BCR-ABL1 DNA the expression of e13a2 BCR-ABL1 mRNA was lower than that of e14a2, an observation that requires confirmation. DNA methods were more sensitive: following the achievement of UMRD by RQ-PCR patients could, on average, be monitored by DNA Q-PCR for an additional 5 months.
Session topic: 8. Chronic myeloid leukemia - Clinical
Keyword(s): BCR-ABL, Molecular response, Minimal residual disease (MRD), Chronic myeloid leukemia
{{ help_message }}
{{filter}}


