
Contributions
Abstract: S420
Type: Oral Presentation
Presentation during EHA22: On Saturday, June 24, 2017 from 12:15 - 12:30
Location: Hall C
Background
JAK2 V617F is the most common somatic mutation in the classical myeloproliferative neoplasms (MPNs) and is also frequent amongst healthy individuals with age-related clonal haemopoiesis (ARCH).
Aims
To investigate the pre-clinical clonal evolution of MPNs.
Methods
We identified 12 individuals with JAK2 V617F mutant MPN from whom blood DNA was available from the time of MPN diagnosis and also from an earlier time point of between 4.5-15.2 years previously (median 10.2 years) when blood was donated for registration to the Cyprus Bone Marrow Donor Registry. We used deep DNA sequencing to interrogate all 24 samples at 15 myeloid mutation hotspots including JAK2 V617, using an established multiplex PCR/MiSeq sequencing protocol that reliably detects nucleotide substitutions present at a variant allele fraction (VAF) ≥0.008. Additionally, for 12 samples with sufficient DNA available, we performed targeted DNA capture for all exons of 41 genes recurrently mutated in myeloid neoplasms using a custom RNA-bait library followed by sequencing on Illumina HiSeq 2500. Finally, we genotyped archival Registry samples for the rs12343867 single nucleotide polymorphism (SNP) (C/T) linked to the JAK2 46/1 haplotype.
Results
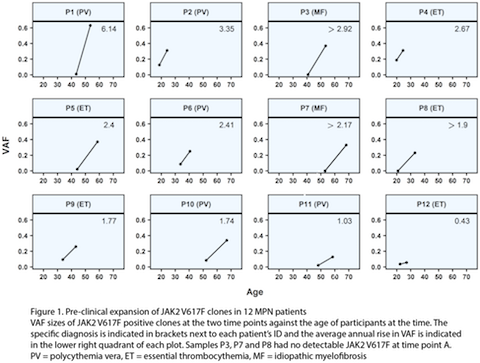
Amplicon sequencing returned a median coverage of 6641 reads per nucleotide (nt) at the studied hotspots. This confirmed the presence of JAK2 V617F in all 12 diagnostic and 9 of 12 archival samples. The remaining 3 samples were JAK2 V617F negative at the sensitivity of our assay (VAF≥0.008). The only other hotspot mutation identified was SRSF2 P95R in one patient, P3, whom had a diagnosis of myelofibrosis. Pulldown sequencing of all exons of 41 genes from 12 samples with sufficient DNA returned an average coverage of 1978 reads per nt and showed a close correlation in JAK2 V617F and SRSF2 P95R VAF quantitations with amplicon sequencing. The JAK2 V617F VAF at MPN diagnosis differed between patients as expected, however the average rate of clonal growth also varied widely between individuals, ranging from 0.36 to 6.2% per annum (Figure 1). Targeted exon capture from 12 of 24 samples, only identified one co-mutation with a VAF >0.02, the SRSF2 P95R in patient P3. As this locus was also amplified by amplicon sequencing, we were able to quantify the SRSF2 P95R VAF in both the diagnostic and the archival DNA sample taken 12.6 years earlier. In the P3 diagnostic sample the VAFs for JAK2 V617F and SRSF2 P95R were similar (0.37 and 0.41 respectively) indicating that they co-occurred in most cells of the neoplastic clone. In the archival sample from P3, the SRSF2 P95R was detectable at a VAF of 0.06, however the JAK2 V617F was absent/undetectable at the sensitivity of our method (VAF≥0.008) indicating the SRSF2 P95R was the clone-founding mutation in this neoplasm. The genotyping results for the rs12343867 SNP revealed a tentative association in our small cohort between homozygosity for the risk allele (C) linked to the JAK2 46/1 haplotype and the average annual increase in JAK2 V617F VAF. This will need to be verified in larger studies.
Conclusion
Our findings reveal that JAK2 V617F neoplasms develop from clonal haematopoiesis over many years.
The rate of clonal expansion of JAK2 V617F clones in the pre-clinical phase was highly variable and although it was tentatively associated with the 46/1 haplotype, the high variability observed suggests that other factors likely influence clonal progression.
Session topic: 15. Myeloproliferative neoplasms - Biology
Keyword(s): mutation analysis, Clonal expansion, Myeloproliferative disorder, Myeloid malignancies
Abstract: S420
Type: Oral Presentation
Presentation during EHA22: On Saturday, June 24, 2017 from 12:15 - 12:30
Location: Hall C
Background
JAK2 V617F is the most common somatic mutation in the classical myeloproliferative neoplasms (MPNs) and is also frequent amongst healthy individuals with age-related clonal haemopoiesis (ARCH).
Aims
To investigate the pre-clinical clonal evolution of MPNs.
Methods
We identified 12 individuals with JAK2 V617F mutant MPN from whom blood DNA was available from the time of MPN diagnosis and also from an earlier time point of between 4.5-15.2 years previously (median 10.2 years) when blood was donated for registration to the Cyprus Bone Marrow Donor Registry. We used deep DNA sequencing to interrogate all 24 samples at 15 myeloid mutation hotspots including JAK2 V617, using an established multiplex PCR/MiSeq sequencing protocol that reliably detects nucleotide substitutions present at a variant allele fraction (VAF) ≥0.008. Additionally, for 12 samples with sufficient DNA available, we performed targeted DNA capture for all exons of 41 genes recurrently mutated in myeloid neoplasms using a custom RNA-bait library followed by sequencing on Illumina HiSeq 2500. Finally, we genotyped archival Registry samples for the rs12343867 single nucleotide polymorphism (SNP) (C/T) linked to the JAK2 46/1 haplotype.
Results
Amplicon sequencing returned a median coverage of 6641 reads per nucleotide (nt) at the studied hotspots. This confirmed the presence of JAK2 V617F in all 12 diagnostic and 9 of 12 archival samples. The remaining 3 samples were JAK2 V617F negative at the sensitivity of our assay (VAF≥0.008). The only other hotspot mutation identified was SRSF2 P95R in one patient, P3, whom had a diagnosis of myelofibrosis. Pulldown sequencing of all exons of 41 genes from 12 samples with sufficient DNA returned an average coverage of 1978 reads per nt and showed a close correlation in JAK2 V617F and SRSF2 P95R VAF quantitations with amplicon sequencing. The JAK2 V617F VAF at MPN diagnosis differed between patients as expected, however the average rate of clonal growth also varied widely between individuals, ranging from 0.36 to 6.2% per annum (Figure 1). Targeted exon capture from 12 of 24 samples, only identified one co-mutation with a VAF >0.02, the SRSF2 P95R in patient P3. As this locus was also amplified by amplicon sequencing, we were able to quantify the SRSF2 P95R VAF in both the diagnostic and the archival DNA sample taken 12.6 years earlier. In the P3 diagnostic sample the VAFs for JAK2 V617F and SRSF2 P95R were similar (0.37 and 0.41 respectively) indicating that they co-occurred in most cells of the neoplastic clone. In the archival sample from P3, the SRSF2 P95R was detectable at a VAF of 0.06, however the JAK2 V617F was absent/undetectable at the sensitivity of our method (VAF≥0.008) indicating the SRSF2 P95R was the clone-founding mutation in this neoplasm. The genotyping results for the rs12343867 SNP revealed a tentative association in our small cohort between homozygosity for the risk allele (C) linked to the JAK2 46/1 haplotype and the average annual increase in JAK2 V617F VAF. This will need to be verified in larger studies.

Conclusion
Our findings reveal that JAK2 V617F neoplasms develop from clonal haematopoiesis over many years.
The rate of clonal expansion of JAK2 V617F clones in the pre-clinical phase was highly variable and although it was tentatively associated with the 46/1 haplotype, the high variability observed suggests that other factors likely influence clonal progression.
Session topic: 15. Myeloproliferative neoplasms - Biology
Keyword(s): mutation analysis, Clonal expansion, Myeloproliferative disorder, Myeloid malignancies


