
Contributions
Abstract: PB2035
Type: Publication Only
Background
Somatic mutations in codons 533-547 of JAK2 exon 12 are highly specific to confirm the diagnosis of polycythemia vera (PV). We have previously proposed techniques for the detection and quantification of JAK2 exon 12 allele burden using a pyrosequencing method (Subbotina T et al, Haematologica 2014). However, due to the high cost of sequencing, developing a two-stage algorithm for detect mutations in JAK2 exon 12 using inexpensive screening is of immediate practically necessity.
Aims
Methods
274 patients with PV or unclear erythrocytosis and with a low JAK2V617F allele burden or unmutated JAK2V617 (51 women, mean age 52.2±15.7 years and 223 men, mean age 43.6±15.6 years) were included in this study. The informed consents from these patients were obtained. The PCR with the additional stage of formation heteroduplexes was performed using the Real-time PCR kit (Syntol, Russia) and CFX 96 Real Time System (Biorad, USA). PCR products were analyzed by electrophoresis in 8% PAGE. The presence of the mutations was identified and confirmed by pyrosequencing method with PyroMark Q24 (Qiagen, Germany). To verify the presence of mutations, the DNA sequences extracted from the clinical samples were cloned into pGem-T vector using standard protocol («Promega», USA), and obtained clones were sequenced using reagents and equipment of the «Applied Biosystems» (USA). JAK2 exon 12 varianceMUT was calculated as a measure of relative changes in allele burden between the baseline and follow-up sample (Theocharides A et al, Haematologica, 2008).
Results
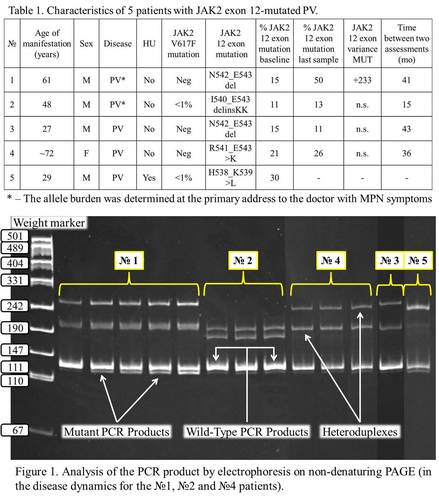
Conclusion
Session topic: 16. Myeloproliferative neoplasms - Clinical
Keyword(s): Myeloproliferative disorder
Abstract: PB2035
Type: Publication Only
Background
Somatic mutations in codons 533-547 of JAK2 exon 12 are highly specific to confirm the diagnosis of polycythemia vera (PV). We have previously proposed techniques for the detection and quantification of JAK2 exon 12 allele burden using a pyrosequencing method (Subbotina T et al, Haematologica 2014). However, due to the high cost of sequencing, developing a two-stage algorithm for detect mutations in JAK2 exon 12 using inexpensive screening is of immediate practically necessity.
Aims
Methods
274 patients with PV or unclear erythrocytosis and with a low JAK2V617F allele burden or unmutated JAK2V617 (51 women, mean age 52.2±15.7 years and 223 men, mean age 43.6±15.6 years) were included in this study. The informed consents from these patients were obtained. The PCR with the additional stage of formation heteroduplexes was performed using the Real-time PCR kit (Syntol, Russia) and CFX 96 Real Time System (Biorad, USA). PCR products were analyzed by electrophoresis in 8% PAGE. The presence of the mutations was identified and confirmed by pyrosequencing method with PyroMark Q24 (Qiagen, Germany). To verify the presence of mutations, the DNA sequences extracted from the clinical samples were cloned into pGem-T vector using standard protocol («Promega», USA), and obtained clones were sequenced using reagents and equipment of the «Applied Biosystems» (USA). JAK2 exon 12 varianceMUT was calculated as a measure of relative changes in allele burden between the baseline and follow-up sample (Theocharides A et al, Haematologica, 2008).
Results

Conclusion
Session topic: 16. Myeloproliferative neoplasms - Clinical
Keyword(s): Myeloproliferative disorder


