DISSECTING RESISTANCE MECHANISMS IN CHRONIC LYMPHOCYTIC LEUKEMIA USING WHOLE-EXOME SEQUENCING: IMPACT OF RECURRENT RPS15 MUTATIONS ON P53 DYSREGULATION
(Abstract release date: 05/21/15)
EHA Library. Ljungström V. 06/12/15; 103153; S121
Disclosure(s): Uppsala UniversityDept of Immunology, Genetics and Pathology

Viktor Ljungström
Contributions
Contributions
Abstract
Abstract: S121
Type: Oral Presentation + travel grant
Presentation during EHA20: From 12.06.2015 11:30 to 12.06.2015 11:45
Location: Room A8
Background
Fludarabine, cyclophosphamide and rituximab (FCR) is the gold-standard first-line regimen in medically fit patients with chronic lymphocytic leukemia (CLL); however, despite good response rates most patients will eventually relapse. Besides TP53 aberrations, the mechanisms leading to relapse after FCR treatment are currently poorly understood.
Aims
To characterize the genetic mechanisms underlying relapse following treatment with FCR using whole-exome sequencing (WES).
Methods
Forty-one CLL patients receiving FCR with either a partial response (PR, with ≥4 cycles of treatment completed) or a complete response (CR, ≥1 cycle of treatment completed) were selected. Pre-treatment and relapse samples (mean time to relapse 3.2 years, range 0.7 – 10.9), together with matched germline DNA for 28 patients, were analyzed by WES. Well-established bioinformatics tools and pipelines were used to process raw sequencing reads, enabling the identification of somatic mutations and also facilitating the analysis of copy-number aberrations (CNA) and absolute cancer cell fractions (CCF).
Results
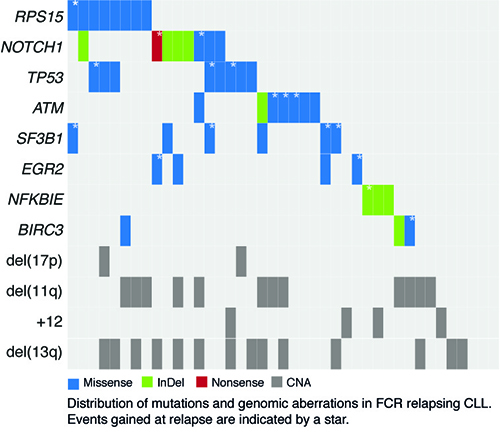
Amongst the 28 patients with matched germline DNA, 1191 somatic variants (>10% allele frequency) were found in the pre-treatment samples and 1334 in the relapse samples, with an average of 15.2 (range, 3-24) and 17.6 (range, 2-32) non-silent mutations per case, respectively. Mutations were predominantly missense substitutions (81%) and less frequently frameshift or in-frame insertions/deletions (14%) or nonsense mutations (5%). As expected, at relapse, a high proportion of cases harbored mutations in genes previously linked to adverse prognosis in CLL: TP53 (n=8; 19.5%), NOTCH1 (n=8; 19.5%), ATM (n=7; 17%), SF3B1 (n=6; 14.6%), NFKBIE (n=4; 9.8%), EGR2 (n=4; 9.8%) and BIRC3 (n=3; 7.3%). Intriguingly, a large proportion of cases also harbored mutations in RPS15 (n=8; 19.5%), a gene encoding a component of the 40S ribosomal subunit. High allele frequencies were observed for RPS15 mutations at both time points (range, 29% - 56%), and all mutations were missense variants residing within a 7 amino-acid evolutionarily conserved region. Besides its role in protein translation, RPS15 has been shown to stabilize p53 by interfering with the MDM2-p53-MDMX network and inhibiting MDM2-mediated p53 degradation. Characterization of two recurrent RPS15 mutations in the HCT116 colorectal cancer cell line transiently expressing either wild-type (wt) or mutant RPS15 revealed impaired ability of RPS15P131S and RPS15G132A in regulating endogenous p53. As both mutations map within the region that interacts with MDM2, this finding strongly suggests that binding of RPS15P131S and RPS15G132A to MDM2 is less efficient compared to wt protein thus leading to more pronounced p53 degradation. Finally, by calculating the absolute CCF for all mutations at both time points allowed monitoring of clonal heterogeneity over time. All 24 cases with available exome-derived CNA data showed mutations expanding ≥0.3 in CCF between the time points (mean 7.4 mutations, range 1-21). Among recurrently mutated genes, i) RPS15 remained stable over time, ii) TP53, EGR2, NOTCH1 and BIRC3 mutations expanded or remained stable, and iii) for SF3B1 and ATM mutations both increasing and decreasing CCFs were observed.
Summary
We provide novel insights into the heterogeneous genetic landscape of CLL relapsing after FCR treatment with our most prominent finding being recurrent RPS15 mutations (19.5%) and with in vitro studies of RPS15 mutations pointing to a novel mechanism for p53 dysregulation in CLL.
Keyword(s): Chronic lymphocytic leukemia, Mutation analysis
Session topic: CLL - Biology: Interacting determinants of CLL ontogeny and evolution
Type: Oral Presentation + travel grant
Presentation during EHA20: From 12.06.2015 11:30 to 12.06.2015 11:45
Location: Room A8
Background
Fludarabine, cyclophosphamide and rituximab (FCR) is the gold-standard first-line regimen in medically fit patients with chronic lymphocytic leukemia (CLL); however, despite good response rates most patients will eventually relapse. Besides TP53 aberrations, the mechanisms leading to relapse after FCR treatment are currently poorly understood.
Aims
To characterize the genetic mechanisms underlying relapse following treatment with FCR using whole-exome sequencing (WES).
Methods
Forty-one CLL patients receiving FCR with either a partial response (PR, with ≥4 cycles of treatment completed) or a complete response (CR, ≥1 cycle of treatment completed) were selected. Pre-treatment and relapse samples (mean time to relapse 3.2 years, range 0.7 – 10.9), together with matched germline DNA for 28 patients, were analyzed by WES. Well-established bioinformatics tools and pipelines were used to process raw sequencing reads, enabling the identification of somatic mutations and also facilitating the analysis of copy-number aberrations (CNA) and absolute cancer cell fractions (CCF).
Results
Amongst the 28 patients with matched germline DNA, 1191 somatic variants (>10% allele frequency) were found in the pre-treatment samples and 1334 in the relapse samples, with an average of 15.2 (range, 3-24) and 17.6 (range, 2-32) non-silent mutations per case, respectively. Mutations were predominantly missense substitutions (81%) and less frequently frameshift or in-frame insertions/deletions (14%) or nonsense mutations (5%). As expected, at relapse, a high proportion of cases harbored mutations in genes previously linked to adverse prognosis in CLL: TP53 (n=8; 19.5%), NOTCH1 (n=8; 19.5%), ATM (n=7; 17%), SF3B1 (n=6; 14.6%), NFKBIE (n=4; 9.8%), EGR2 (n=4; 9.8%) and BIRC3 (n=3; 7.3%). Intriguingly, a large proportion of cases also harbored mutations in RPS15 (n=8; 19.5%), a gene encoding a component of the 40S ribosomal subunit. High allele frequencies were observed for RPS15 mutations at both time points (range, 29% - 56%), and all mutations were missense variants residing within a 7 amino-acid evolutionarily conserved region. Besides its role in protein translation, RPS15 has been shown to stabilize p53 by interfering with the MDM2-p53-MDMX network and inhibiting MDM2-mediated p53 degradation. Characterization of two recurrent RPS15 mutations in the HCT116 colorectal cancer cell line transiently expressing either wild-type (wt) or mutant RPS15 revealed impaired ability of RPS15P131S and RPS15G132A in regulating endogenous p53. As both mutations map within the region that interacts with MDM2, this finding strongly suggests that binding of RPS15P131S and RPS15G132A to MDM2 is less efficient compared to wt protein thus leading to more pronounced p53 degradation. Finally, by calculating the absolute CCF for all mutations at both time points allowed monitoring of clonal heterogeneity over time. All 24 cases with available exome-derived CNA data showed mutations expanding ≥0.3 in CCF between the time points (mean 7.4 mutations, range 1-21). Among recurrently mutated genes, i) RPS15 remained stable over time, ii) TP53, EGR2, NOTCH1 and BIRC3 mutations expanded or remained stable, and iii) for SF3B1 and ATM mutations both increasing and decreasing CCFs were observed.
Summary
We provide novel insights into the heterogeneous genetic landscape of CLL relapsing after FCR treatment with our most prominent finding being recurrent RPS15 mutations (19.5%) and with in vitro studies of RPS15 mutations pointing to a novel mechanism for p53 dysregulation in CLL.
Keyword(s): Chronic lymphocytic leukemia, Mutation analysis

Session topic: CLL - Biology: Interacting determinants of CLL ontogeny and evolution
Abstract: S121
Type: Oral Presentation + travel grant
Presentation during EHA20: From 12.06.2015 11:30 to 12.06.2015 11:45
Location: Room A8
Background
Fludarabine, cyclophosphamide and rituximab (FCR) is the gold-standard first-line regimen in medically fit patients with chronic lymphocytic leukemia (CLL); however, despite good response rates most patients will eventually relapse. Besides TP53 aberrations, the mechanisms leading to relapse after FCR treatment are currently poorly understood.
Aims
To characterize the genetic mechanisms underlying relapse following treatment with FCR using whole-exome sequencing (WES).
Methods
Forty-one CLL patients receiving FCR with either a partial response (PR, with ≥4 cycles of treatment completed) or a complete response (CR, ≥1 cycle of treatment completed) were selected. Pre-treatment and relapse samples (mean time to relapse 3.2 years, range 0.7 – 10.9), together with matched germline DNA for 28 patients, were analyzed by WES. Well-established bioinformatics tools and pipelines were used to process raw sequencing reads, enabling the identification of somatic mutations and also facilitating the analysis of copy-number aberrations (CNA) and absolute cancer cell fractions (CCF).
Results
Amongst the 28 patients with matched germline DNA, 1191 somatic variants (>10% allele frequency) were found in the pre-treatment samples and 1334 in the relapse samples, with an average of 15.2 (range, 3-24) and 17.6 (range, 2-32) non-silent mutations per case, respectively. Mutations were predominantly missense substitutions (81%) and less frequently frameshift or in-frame insertions/deletions (14%) or nonsense mutations (5%). As expected, at relapse, a high proportion of cases harbored mutations in genes previously linked to adverse prognosis in CLL: TP53 (n=8; 19.5%), NOTCH1 (n=8; 19.5%), ATM (n=7; 17%), SF3B1 (n=6; 14.6%), NFKBIE (n=4; 9.8%), EGR2 (n=4; 9.8%) and BIRC3 (n=3; 7.3%). Intriguingly, a large proportion of cases also harbored mutations in RPS15 (n=8; 19.5%), a gene encoding a component of the 40S ribosomal subunit. High allele frequencies were observed for RPS15 mutations at both time points (range, 29% - 56%), and all mutations were missense variants residing within a 7 amino-acid evolutionarily conserved region. Besides its role in protein translation, RPS15 has been shown to stabilize p53 by interfering with the MDM2-p53-MDMX network and inhibiting MDM2-mediated p53 degradation. Characterization of two recurrent RPS15 mutations in the HCT116 colorectal cancer cell line transiently expressing either wild-type (wt) or mutant RPS15 revealed impaired ability of RPS15P131S and RPS15G132A in regulating endogenous p53. As both mutations map within the region that interacts with MDM2, this finding strongly suggests that binding of RPS15P131S and RPS15G132A to MDM2 is less efficient compared to wt protein thus leading to more pronounced p53 degradation. Finally, by calculating the absolute CCF for all mutations at both time points allowed monitoring of clonal heterogeneity over time. All 24 cases with available exome-derived CNA data showed mutations expanding ≥0.3 in CCF between the time points (mean 7.4 mutations, range 1-21). Among recurrently mutated genes, i) RPS15 remained stable over time, ii) TP53, EGR2, NOTCH1 and BIRC3 mutations expanded or remained stable, and iii) for SF3B1 and ATM mutations both increasing and decreasing CCFs were observed.
Summary
We provide novel insights into the heterogeneous genetic landscape of CLL relapsing after FCR treatment with our most prominent finding being recurrent RPS15 mutations (19.5%) and with in vitro studies of RPS15 mutations pointing to a novel mechanism for p53 dysregulation in CLL.
Keyword(s): Chronic lymphocytic leukemia, Mutation analysis

Session topic: CLL - Biology: Interacting determinants of CLL ontogeny and evolution
Type: Oral Presentation + travel grant
Presentation during EHA20: From 12.06.2015 11:30 to 12.06.2015 11:45
Location: Room A8
Background
Fludarabine, cyclophosphamide and rituximab (FCR) is the gold-standard first-line regimen in medically fit patients with chronic lymphocytic leukemia (CLL); however, despite good response rates most patients will eventually relapse. Besides TP53 aberrations, the mechanisms leading to relapse after FCR treatment are currently poorly understood.
Aims
To characterize the genetic mechanisms underlying relapse following treatment with FCR using whole-exome sequencing (WES).
Methods
Forty-one CLL patients receiving FCR with either a partial response (PR, with ≥4 cycles of treatment completed) or a complete response (CR, ≥1 cycle of treatment completed) were selected. Pre-treatment and relapse samples (mean time to relapse 3.2 years, range 0.7 – 10.9), together with matched germline DNA for 28 patients, were analyzed by WES. Well-established bioinformatics tools and pipelines were used to process raw sequencing reads, enabling the identification of somatic mutations and also facilitating the analysis of copy-number aberrations (CNA) and absolute cancer cell fractions (CCF).
Results
Amongst the 28 patients with matched germline DNA, 1191 somatic variants (>10% allele frequency) were found in the pre-treatment samples and 1334 in the relapse samples, with an average of 15.2 (range, 3-24) and 17.6 (range, 2-32) non-silent mutations per case, respectively. Mutations were predominantly missense substitutions (81%) and less frequently frameshift or in-frame insertions/deletions (14%) or nonsense mutations (5%). As expected, at relapse, a high proportion of cases harbored mutations in genes previously linked to adverse prognosis in CLL: TP53 (n=8; 19.5%), NOTCH1 (n=8; 19.5%), ATM (n=7; 17%), SF3B1 (n=6; 14.6%), NFKBIE (n=4; 9.8%), EGR2 (n=4; 9.8%) and BIRC3 (n=3; 7.3%). Intriguingly, a large proportion of cases also harbored mutations in RPS15 (n=8; 19.5%), a gene encoding a component of the 40S ribosomal subunit. High allele frequencies were observed for RPS15 mutations at both time points (range, 29% - 56%), and all mutations were missense variants residing within a 7 amino-acid evolutionarily conserved region. Besides its role in protein translation, RPS15 has been shown to stabilize p53 by interfering with the MDM2-p53-MDMX network and inhibiting MDM2-mediated p53 degradation. Characterization of two recurrent RPS15 mutations in the HCT116 colorectal cancer cell line transiently expressing either wild-type (wt) or mutant RPS15 revealed impaired ability of RPS15P131S and RPS15G132A in regulating endogenous p53. As both mutations map within the region that interacts with MDM2, this finding strongly suggests that binding of RPS15P131S and RPS15G132A to MDM2 is less efficient compared to wt protein thus leading to more pronounced p53 degradation. Finally, by calculating the absolute CCF for all mutations at both time points allowed monitoring of clonal heterogeneity over time. All 24 cases with available exome-derived CNA data showed mutations expanding ≥0.3 in CCF between the time points (mean 7.4 mutations, range 1-21). Among recurrently mutated genes, i) RPS15 remained stable over time, ii) TP53, EGR2, NOTCH1 and BIRC3 mutations expanded or remained stable, and iii) for SF3B1 and ATM mutations both increasing and decreasing CCFs were observed.
Summary
We provide novel insights into the heterogeneous genetic landscape of CLL relapsing after FCR treatment with our most prominent finding being recurrent RPS15 mutations (19.5%) and with in vitro studies of RPS15 mutations pointing to a novel mechanism for p53 dysregulation in CLL.
Keyword(s): Chronic lymphocytic leukemia, Mutation analysis

Session topic: CLL - Biology: Interacting determinants of CLL ontogeny and evolution
{{ help_message }}
{{filter}}


