CLIP, Department of Pediatric Haematology and Oncology

Contributions
Type: Oral Presentation
Presentation during EHA20: From 13.06.2015 11:45 to 13.06.2015 12:00
Location: Room C1
Background
Minimal residual disease (MRD) monitoring using antigen receptor-based quantitative PCR (qPCR) became a gold standard in the management of acute lymphoblastic leukemia (ALL), despite being technically and financially demanding. MRD detection based on next generation sequencing (NGS) of antigen receptor genes rearrangements allows for a highly specific and sensitive detection of MRD without the need for labourious optimization of patient-specific assays.
Aims
To establish MRD detection by NGS of immunoglobulin heavy chain (IGH) rearrangements and compare MRD levels at BFM-protocols stratification timepoints with qPCR and flow cytometry (FC).
Methods
The libraries for sequencing were prepared from 450ng of diagnostic bone marrow DNA and 50ng of polyclonal DNA. Two-round PCR was used for library preparation: in the 1st round the IGH rearrangements were amplified with Biomed-2 FR3 primers. In the 2nd round of PCR sequencing adaptors and barcodes were attached. Libraries were sequenced on Ion Torrent PGM/Ion Proton sequencers.
For detection of reads containing the clonal sequence from diagnosis we used our own bioinformatics algorithm.
Results
We sequenced 213 samples from 63 patients with childhood ALL treated according to the AIEOP-BFM-ALL 2000 protocol and 14 more patients with relapse from previous and current frontline treatment protocols with the median coverage 719,904 reads per sample . Eighty-four (39.4%) samples were negative by both methods. Sixteen (7.5%) samples were positive by NGS and negative by qPCR, and 16 (7.5%) samples were positive by qPCR and negative by NGS. This caused a shift in risk group stratification in 30% of patients, mainly between standard risk and intermediate risk group patients. The overall correlation of both methods was good (R2=0.71).
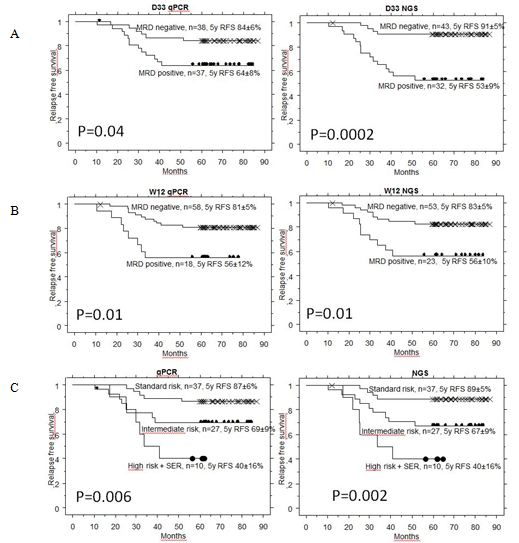
NGS approach detected significantly higher MRD than qPCR at day 33 in patients who later relapsed (p=0.001). NGS-MRD positivity at day 33 seems to provide a more accurate prediction of relapse than qPCR-MRD positivity (Fig. 1A). At day 78, the predictive value of NGS was comparable to qPCR (Fig. 1B). Combined day 33 and 78 MRD used for defining of SR, IR and HR (+SER) groups on BFM trials gave again similar results as qPCR (Fig. 1C). Similarly to FC, low NGS MRD defined a group with excellent prognosis at day 15.
Summary
NGS will speed up the process of MRD detection and provide results at early time points, which is currently not possible due to the long time needed for qPCR preparations. The correlation of NGS and qPCR was good with the majority of the differrences below the reproducible sensitivity of methods, which caused shifts mainly between SR and IR groups.
We showed that day 33 NGS MRD levels were higher than qPCR MRD levels in patients who subsequently relapsed, which was reflected in a slightly better prediction of relapse based on d33 NGS. The outcome of patients stratified into risk groups by combined d33/d78 MRD was similar for NGS and qPCR, despite the 30% of patients being differently stratified by NGS.
At present, the main drawback of the Ig/TCR-exploring NGS methods is the lack of standardization both in the experimental setting and in data analysis. Therefore, the European network “EuroClonality NGS Consortium”, has been formed to optimize and standardize NGS workflow. .Before NGS based MRD can safely replace qPCR for treatment guiding in ALL, robust results on comparability of both methods by standardized methodology should be validated within prospective clinical trials.
Keyword(s): Acute lymphoblastic leukemia, Minimal residual disease (MRD)
Session topic: Translational studies in ALL
Type: Oral Presentation
Presentation during EHA20: From 13.06.2015 11:45 to 13.06.2015 12:00
Location: Room C1
Background
Minimal residual disease (MRD) monitoring using antigen receptor-based quantitative PCR (qPCR) became a gold standard in the management of acute lymphoblastic leukemia (ALL), despite being technically and financially demanding. MRD detection based on next generation sequencing (NGS) of antigen receptor genes rearrangements allows for a highly specific and sensitive detection of MRD without the need for labourious optimization of patient-specific assays.
Aims
To establish MRD detection by NGS of immunoglobulin heavy chain (IGH) rearrangements and compare MRD levels at BFM-protocols stratification timepoints with qPCR and flow cytometry (FC).
Methods
The libraries for sequencing were prepared from 450ng of diagnostic bone marrow DNA and 50ng of polyclonal DNA. Two-round PCR was used for library preparation: in the 1st round the IGH rearrangements were amplified with Biomed-2 FR3 primers. In the 2nd round of PCR sequencing adaptors and barcodes were attached. Libraries were sequenced on Ion Torrent PGM/Ion Proton sequencers.
For detection of reads containing the clonal sequence from diagnosis we used our own bioinformatics algorithm.
Results
We sequenced 213 samples from 63 patients with childhood ALL treated according to the AIEOP-BFM-ALL 2000 protocol and 14 more patients with relapse from previous and current frontline treatment protocols with the median coverage 719,904 reads per sample . Eighty-four (39.4%) samples were negative by both methods. Sixteen (7.5%) samples were positive by NGS and negative by qPCR, and 16 (7.5%) samples were positive by qPCR and negative by NGS. This caused a shift in risk group stratification in 30% of patients, mainly between standard risk and intermediate risk group patients. The overall correlation of both methods was good (R2=0.71).
NGS approach detected significantly higher MRD than qPCR at day 33 in patients who later relapsed (p=0.001). NGS-MRD positivity at day 33 seems to provide a more accurate prediction of relapse than qPCR-MRD positivity (Fig. 1A). At day 78, the predictive value of NGS was comparable to qPCR (Fig. 1B). Combined day 33 and 78 MRD used for defining of SR, IR and HR (+SER) groups on BFM trials gave again similar results as qPCR (Fig. 1C). Similarly to FC, low NGS MRD defined a group with excellent prognosis at day 15.
Summary
NGS will speed up the process of MRD detection and provide results at early time points, which is currently not possible due to the long time needed for qPCR preparations. The correlation of NGS and qPCR was good with the majority of the differrences below the reproducible sensitivity of methods, which caused shifts mainly between SR and IR groups.
We showed that day 33 NGS MRD levels were higher than qPCR MRD levels in patients who subsequently relapsed, which was reflected in a slightly better prediction of relapse based on d33 NGS. The outcome of patients stratified into risk groups by combined d33/d78 MRD was similar for NGS and qPCR, despite the 30% of patients being differently stratified by NGS.
At present, the main drawback of the Ig/TCR-exploring NGS methods is the lack of standardization both in the experimental setting and in data analysis. Therefore, the European network “EuroClonality NGS Consortium”, has been formed to optimize and standardize NGS workflow. .Before NGS based MRD can safely replace qPCR for treatment guiding in ALL, robust results on comparability of both methods by standardized methodology should be validated within prospective clinical trials.
Keyword(s): Acute lymphoblastic leukemia, Minimal residual disease (MRD)

Session topic: Translational studies in ALL


